| 规格 | 价格 | 库存 | 数量 |
|---|---|---|---|
| 1mg |
|
||
| 5mg |
|
||
| 10mg |
|
||
| 25mg |
|
||
| 50mg |
|
||
| 100mg |
|
||
| 250mg |
|
||
| 500mg |
|
||
| Other Sizes |
|
| 靶点 |
Ionizable cationic lipid; mRNA vaccine delivery
|
|---|---|
| 体外研究 (In Vitro) |
迄今为止,三种可电离的阳离子脂质已被批准用于基于RNA的治疗中的临床用途:DLin-MC3-DMA(十七烷-6,9,28,31-四烯-19-基-4-(二甲氨基)丁酸酯)、ALC-0315(4-羟基丁基)氮杂二基)双(己烷-6,1-二烷基)双(2-己基癸酸酯)和SM-102(十七烷-9-基8-((2-羟乙基)(6-氧代-6-(十一烷氧基)己基)氨基)辛酸酯)。MC3被设计用于将siRNA递送到肝细胞,以治疗遗传性转甲状腺素蛋白淀粉样变性(hATTR)。ALC-0315和SM-102是专为mRNA递送而设计的,因为它们是辉瑞/BioNTech/Acuitas和莫德纳分别开发的基于mRNA的严重急性呼吸系统综合征冠状病毒2型疫苗中的可电离阳离子脂质。17,18这三种药物制剂都使用类似的“辅助”脂质和脂质摩尔比,约50%的可电离离子脂质,10%的DSPC(1,2-二硬脂酰-sn-甘油-3-磷酸胆碱),38.5%的胆固醇和1.5%的PEG-DMG(1,2-二肉豆蔻酰rac-甘油-3-甲氧基聚乙二醇-2000)。与ALC-0315和SM-102相比,MC3具有主要的结构差异,因此可能存在效力和毒性差异。在这里,我们报告了含有MC3和ALC-0315的LNPs的直接比较,比较了它们的肝毒性,以及在体内将siRNA货物输送到肝细胞和HSC的能力。由于与ALC-0315的结构相似,本研究未对SM-102进行研究;两种可电离的脂质都表现出相似的支化和相同的官能团(一个羟基、一个叔胺、两个酯和仅饱和烃)[2]。
|
| 体内研究 (In Vivo) |
在这项研究中,研究人员评估了基于SM-102的脂质纳米粒(LNPs)体内共递送Cas9 mRNA和引导RNA(gRNA)对小鼠和更高物种中HBV cccDNA和整合DNA的影响。CRISPR纳米粒子治疗使AAV-HBV1.04转导的小鼠肝脏中HBcAg、HBsAg和cccDNA的水平分别降低了53%、73%和64%。在HBV感染的树鼩中,该治疗实现了病毒RNA减少70%和cccDNA减少35%。在HBV转基因小鼠中,观察到90%的HBV RNA抑制和95%的DNA抑制。CRISPR纳米粒子治疗在小鼠和树鼩中都具有良好的耐受性,因为没有观察到肝酶升高和最小脱靶。该研究表明,基于SM-102的CRISPR在体内靶向HBV游离和整合DNA方面是安全有效的。基于SM-102的LNPs提供的系统可作为治疗HBV感染的潜在策略[4]。
|
| 酶活实验 |
可电离脂质纳米粒中mRNA合成和包封的方案:[3]
基本方案1:通过体外转录和酶盖和拖尾合成mRNA 基本方案2:将mRNA封装到可电离的脂质纳米颗粒中 替代方案:使用预制囊泡对mRNA进行小规模封装 基本方案3:mRNA可电离脂质纳米颗粒的表征和质量控制 有关更多详细信息,请参阅https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/cpz1.898 |
| 细胞实验 |
SM-102(1-辛基壬基8-[(2-羟乙基)[6-氧代-6-(十一烷氧基)己基]氨基]-辛酸酯)是一种氨基阳离子脂质,专为脂质纳米颗粒的形成而设计,是ModernaTM新冠肺炎疫苗中的基本成分之一。然而,它在多大程度上可以改变不同类型的质膜离子电流在很大程度上仍然不确定。在这项研究中,我们研究了SM-102对两种内分泌细胞(如大鼠垂体瘤(GH3)细胞和小鼠间质瘤(MA-10)细胞)或小胶质细胞(BV2)中离子电流的影响。研究了浸泡在高K+、无Ca2+细胞外溶液中的这些细胞中的超极化激活K+电流,以评估SM-102对erg介导的K+电流(IK(erg))的振幅和滞后的影响。SM-102的添加以浓度依赖的方式有效地阻断IK(erg),其半最大浓度(IC50)为108μM,该值与增强电流失活时间常数所需的KD值(即134μM)相似。在SM-102存在的情况下,IK(erg)对长时间等腰三角形斜坡脉冲的滞后强度有效降低。细胞暴露于转染试剂TurboFectinTM 8.0(0.1%,v/v)能够有效抑制超极化激活的IK(erg),并延长电流的失活时间。此外,在用精胺(30μM)透析的GH3细胞中,IK(erg)振幅逐渐降低;此外,SM-102(100μM)或TurboFectin(0.1%)的进一步浴应用进一步降低了电流幅度。在MA-10 Leydig细胞中,IK(erg)也被SM-102或TurboFectin的存在阻断。SM-102诱导的MA-10细胞IK(erg)抑制的IC50值为98μM。在BV2小胶质细胞中,SM-102抑制了内向整流K+电流的振幅。综上所述,假设存在类似的体内发现,SM-102的存在会浓度依赖性地抑制内分泌细胞(如GH3或MA-10细胞)中的IK(erg),并且这种作用可能有助于它们的功能活动[5]。
|
| 动物实验 |
Bioluminescence imaging[4]
To detect the tissue distribution of SM-102-based LNPs formulation containing mRNA upon intravenous administration in mice, bioluminescence imaging (BLI) analysis was performed by using SM-102-based LNPs containing a firefly luciferase (FLuc) reporter mRNA. Briefly, 6–8 weeks old C57BL/6 mice were inoculated with 20 μg SM-102-based LNPs containing FLuc mRNA via the i.v. route. Six hours after injection, animals were given an intraperitoneal injection of luciferase substrate, and fluorescent signals were collected with an IVIS Spectrum instrument. The heart, liver, spleen, lung, and kidney tissues were collected, and the fluorescence signal of each tissue was detected by IVIS Spectrum instrument.[4] Mouse experiments[4] C57BL/6 mice and HBV transgenic mice were used. For AAV-HBV1.04 transduced mouse model, 6–8 weeks C57BL/6 male mice were injected with 5 × 1010 viral genome equivalents/mouse D genotype AAV-HBV1.04 through tail vein. After 10 days, the model was successfully built. To compare the efficiencies of CKK-E12- and SM-102-based nanoparticles, one dose of these two kinds of LNPs (3 mg/kg body weight) were injected into the mice respectively, and mice received PBS as control group. Then blood samples were collected at 2, 4 and 6 days after treatment. Livers were collected at 6 days after therapy. And in D genotype AAV-HBV1.04 transduced mice, we also evaluated SM-102-based LNPs encapsulating Cas9 mRNA and HBV targeting gRNAs effect with different doses. The mice received 3 or 1.5 mg/kg body weight SM-102-based LNPs encapsulating Cas9 mRNA and HBV targeting gRNAs. And mice received PBS as control group. The blood samples were collected at 2, 4 and 6 days after treatment. The livers were collected at 6 days after LNPs treatment. For HBV transgenic mice, 6 weeks old male mice were used to detect the therapeutic effect of LNPs. Two doses of nanoparticles encapsulating Cas9 mRNA with HBV or GFP targeting gRNAs were injected via i.v. into the mice at 1.5 mg/kg body weight. The blood samples were collected at day 2, 4, 11, 18 and 25 after therapy. The mice were sacrificed on day 25 post LNPs treatment. The blood was centrifuged at 1800 rpm for 15 min at 4 °C. Next, we collected the serum obtained to examine HBeAg, HBsAg, ALT and AST. The levels of HBV RNA, DNA and protein in the liver were determined. [4] Tree shrew experiments[4] The tree shrews used in this study were originated from the Kunming Institute of Zoology, Chinese Academy of Science (Yunnan, China). Tacrolimus was dissolved in 5% DMSO, 40% PEG300, 5% Tween-80 and 50% saline. The resulting tacrolimus solution was injected via i.m. into the tree shrews at 0.08 mg/day/kg body weight in a total volume of 50 μl for the first 14 days. Tree shrews were inoculated i.p. with dexamethasone 10 mg/kg 2 days before and after HBV infection. Liposomal alendronate (CAS 121268-17-5) was injected via i.v. into tree shrews at 0.5 mg/kg the day before HBV infection. After Tacrolimus treated for 7 days, tree shrews were infected with HBV DNA positive human serum via the tail vein and the dose was adjusted to 106 copies/tree shrew. Three days later, tree shrews received 106 copies/tree shrew HBV via i.p. again. After the model successfully built, two doses of SM-102-based LNPs were injected into tree shrews at 1.5 mg/kg body weight for therapy. For control group, the tree shrews received LNPs encapsulating Cas9 mRNA and GFP targeting gRNAs. On day 2 and 4 after the treatment, the blood samples were collected. Tree shrews were sacrificed on day 5 post the nanoparticles treatment to determine the level of HBV RNA, DNA, protein and SMC5 in liver. And hepatitis B e antigen (HBeAg), HBsAg and HBV DNA levels in serum were also detected. SM-102-based LNP formulation[4] LNPs were prepared by microfluidic techniques as reported previously (Mol. Ther., 26 (2018), pp. 1509-1519, 10.1016/j.ymthe.2018.03.010). In brief, SM-102 lipids were dissolved in ethanol at molar ratios of 50: 10: 38.5: 1.5 (ionizable lipid: DSPC: cholesterol: PEG-lipid). Chemically modified (2’ O-Methyl RNA/Phosphorothioated) HBV targeting gRNA and GFP targeting gRNA was synthesized. The RNA cargo (1: 1 wt ratio Cas9 mRNA: gRNA) was dissolved in 6.25 mM sodium acetate buffer (pH 5.0). Then, one volume of lipid mixture and three volume of RNA cargo were injected in to a NanoAssemblr microfluidic mixing device. The LNPs were dialyzed against PBS (pH 7.4) for overnight. The LNPs were passed through a 0.22 μm filter, and stored at 4 °C until use. RNA encapsulation efficiency was characterized by Ribogreen assay. The LNP size and ζ-potential were measured using a dynamic light scattering (DLS) technique. |
| 参考文献 |
[1]. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol Ther. 2018 Jun 6;26(6):1509-1519.
[2]. Comparison of DLin-MC3-DMA and ALC-0315 for siRNA Delivery to Hepatocytes and Hepatic Stellate Cells. Mol Pharm. 2022;19(7):2175-2182. [3]. mRNA Synthesis and Encapsulation in Ionizable Lipid Nanoparticles. Curr Protoc. 2023;3(9):e898. [4]. Co-delivery of Cas9 mRNA and guide RNAs edits hepatitis B virus episomal and integration DNA in mouse and tree shrew models. Antiviral Res . 2023 Jul:215:105618. [5]. Effective Perturbations on the Amplitude and Hysteresis of Erg-Mediated Potassium Current Caused by 1-Octylnonyl 8-[(2-hydroxyethyl)[6-oxo-6(undecyloxy)hexyl]amino]-octanoate (SM-102), a Cationic Lipid. Biomedicines . 2021 Oct 1;9(10):1367. |
| 其他信息 |
Refer to Figure 1 for safe stopping points indicated by red arrows.
Basic Protocol 1: Synthesis of mRNA by in vitro transcription and enzymatic capping and tailing Allow 2 to 4 days to complete the entire protocol including the production and assessment of capped and tailed mRNA. Four days will be required if precipitations are planned overnight. The IVT, capping and tailing reactions all take approximately half a day. All reactions can be set up in ∼1 hr followed by the required incubation time of two hours for IVT and one hour for capping/tailing. Precipitation of the mRNA requires a 30 min centrifugation step and resuspension of the RNA takes ∼10 min. Quality assessment of the mRNA by Nanodrop, agarose gel and automated gel electrophoresis takes 1 hr. Note when scaling up, more time is required particularly when multiple or larger mRNA pellets need to be resuspended in nuclease-free water. Basic Protocol 2: Encapsulation of mRNA into iLNPs Preparation of lipid solutions may be carried out in advance of the formulation step. Otherwise, the entire encapsulation protocol must be carried out on the same day. Allow an hour for all the reagents to come to room temperature before use and an hour to carry out the formulation and dilution into DPBS step. The centrifugal concentration step is dependent on the particle size, total sample volume and desired end volume. Typically allow 1 to 4 hr. Once concentrated, the iLNP solution can be stored in the fridge until the dilution requirements are determined by the RiboGreen assay. Alternate Protocol: Small-scale encapsulation of mRNA using preformed vesicles Same as for Basic Protocol 2. Basic Protocol 3: Characterization and quality control of mRNA iLNPs Allow 1 to 2 hr for the RiboGreen assay including allowing the kit to warm to room temperature from the fridge. DLS (size, PDI, zeta potential) should be carried out on the final, diluted sample before it is used for biological evaluation. Preparation of samples takes a few minutes and analysis time varies across different instruments, but it is typically 5 to 10 min per sample. The buffers used in the TNS assay must be at room temperature before use. Depending on the size of the aliquot this may take several hours. Buffers can be moved to the fridge the night before the assay is to be run to reduce the time required to warm the buffers. The assay requires 40 aliquots in a 96-well plate per iLNP sample therefore allow 10 to 15 min per iLNP sample to prepare the plate and 10 min to read the plate. Allow 30 min for the mRNA extraction protocol. See above (Basic Protocol 1) for analysis of extracted mRNA by automated gel electrophoresis.[3] With 296 million chronically infected individuals worldwide, hepatitis B virus (HBV) causes a major health burden. The major challenge to cure HBV infection lies in the fact that the source of persistence infection, viral episomal covalently closed circular DNA (cccDNA), could not be targeted. In addition, HBV DNA integration, although normally results in replication-incompetent transcripts, considered as oncogenic. Though several studies evaluated the potential of gene-editing approaches to target HBV, previous in vivo studies have been of limited relevance to authentic HBV infection, as the models do not contain HBV cccDNA or feature a complete HBV replication cycle under competent host immune system. In this study, we evaluated the effect of in vivo codelivery of Cas9 mRNA and guide RNAs (gRNAs) by SM-102-based lipid nanoparticles (LNPs) on HBV cccDNA and integrated DNA in mouse and a higher species. CRISPR nanoparticle treatment decreased the levels of HBcAg, HBsAg and cccDNA in AAV-HBV1.04 transduced mouse liver by 53%, 73% and 64% respectively. In HBV infected tree shrews, the treatment achieved 70% reduction of viral RNA and 35% reduction of cccDNA. In HBV transgenic mouse, 90% inhibition of HBV RNA and 95% inhibition of DNA were observed. CRISPR nanoparticle treatment was well tolerated in both mouse and tree shrew, as no elevation of liver enzymes and minimal off-target was observed. Our study demonstrated that SM-102-based CRISPR is safe and effective in targeting HBV episomal and integration DNA in vivo. The system delivered by SM-102-based LNPs may be used as a potential therapeutic strategy against HBV infection.[4] |
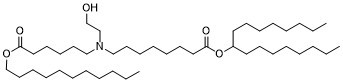
| 分子式 |
C44H87NO5
|
|---|---|
| 分子量 |
710.1653
|
| 精确质量 |
709.66
|
| 元素分析 |
C, 74.42; H, 12.35; N, 1.97; O, 11.26
|
| CAS号 |
2089251-47-6
|
| PubChem CID |
126697616
|
| 外观&性状 |
Colorless to light yellow oily liquid
|
| LogP |
15.5
|
| tPSA |
76.1Ų
|
| 氢键供体(HBD)数目 |
1
|
| 氢键受体(HBA)数目 |
6
|
| 可旋转键数目(RBC) |
43
|
| 重原子数目 |
50
|
| 分子复杂度/Complexity |
686
|
| 定义原子立体中心数目 |
0
|
| InChi Key |
BGNVBNJYBVCBJH-UHFFFAOYSA-N
|
| InChi Code |
InChI=1S/C44H87NO5/c1-4-7-10-13-16-17-18-24-32-41-49-43(47)35-29-25-31-38-45(39-40-46)37-30-23-19-22-28-36-44(48)50-42(33-26-20-14-11-8-5-2)34-27-21-15-12-9-6-3/h42,46H,4-41H2,1-3H3
|
| 化学名 |
heptadecan-9-yl 8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino)octanoate
|
| 别名 |
SM102; SM-102; heptadecan-9-yl 8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino)octanoate; SM-102 (Excipient); T7OBQ65G2I; 1-Octylnonyl 8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino)octanoate; heptadecan-9-yl 8-[2-hydroxyethyl-(6-oxo-6-undecoxyhexyl)amino]octanoate; Heptadecan-9-yl 8-[(2-Hydroxyethyl)[6-oxo-6-(undecyloxy)hexyl]amino]octanoate; SM 102
|
| HS Tariff Code |
2934.99.9001
|
| 存储方式 |
Powder -20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month |
| 运输条件 |
Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs)
|
| 溶解度 (体外实验) |
Ethanol :≥ 100 mg/mL (~140.81 mM)
DMSO : ~100 mg/mL (~140.81 mM) |
|---|---|
| 溶解度 (体内实验) |
配方 1 中的溶解度: ≥ 2.5 mg/mL (3.52 mM) (饱和度未知) in 10% DMSO + 40% PEG300 + 5% Tween80 + 45% Saline (这些助溶剂从左到右依次添加,逐一添加), 澄清溶液。
例如,若需制备1 mL的工作液,可将100 μL 25.0 mg/mL澄清DMSO储备液加入到400 μL PEG300中,混匀;然后向上述溶液中加入50 μL Tween-80,混匀;加入450 μL生理盐水定容至1 mL。 *生理盐水的制备:将 0.9 g 氯化钠溶解在 100 mL ddH₂O中,得到澄清溶液。 配方 2 中的溶解度: 2.5 mg/mL (3.52 mM) in 10% DMSO + 90% (20% SBE-β-CD in Saline) (这些助溶剂从左到右依次添加,逐一添加), 悬浊液; 超声助溶。 例如,若需制备1 mL的工作液,可将 100 μL 25.0 mg/mL澄清DMSO储备液加入900 μL 20% SBE-β-CD生理盐水溶液中,混匀。 *20% SBE-β-CD 生理盐水溶液的制备(4°C,1 周):将 2 g SBE-β-CD 溶解于 10 mL 生理盐水中,得到澄清溶液。 View More
配方 3 中的溶解度: ≥ 2.5 mg/mL (3.52 mM) (饱和度未知) in 10% DMSO + 90% Corn Oil (这些助溶剂从左到右依次添加,逐一添加), 澄清溶液。 1、请先配制澄清的储备液(如:用DMSO配置50 或 100 mg/mL母液(储备液)); 2、取适量母液,按从左到右的顺序依次添加助溶剂,澄清后再加入下一助溶剂。以 下列配方为例说明 (注意此配方只用于说明,并不一定代表此产品 的实际溶解配方): 10% DMSO → 40% PEG300 → 5% Tween-80 → 45% ddH2O (或 saline); 假设最终工作液的体积为 1 mL, 浓度为5 mg/mL: 取 100 μL 50 mg/mL 的澄清 DMSO 储备液加到 400 μL PEG300 中,混合均匀/澄清;向上述体系中加入50 μL Tween-80,混合均匀/澄清;然后继续加入450 μL ddH2O (或 saline)定容至 1 mL; 3、溶剂前显示的百分比是指该溶剂在最终溶液/工作液中的体积所占比例; 4、 如产品在配制过程中出现沉淀/析出,可通过加热(≤50℃)或超声的方式助溶; 5、为保证最佳实验结果,工作液请现配现用! 6、如不确定怎么将母液配置成体内动物实验的工作液,请查看说明书或联系我们; 7、 以上所有助溶剂都可在 Invivochem.cn网站购买。 |
| 制备储备液 | 1 mg | 5 mg | 10 mg | |
| 1 mM | 1.4081 mL | 7.0406 mL | 14.0811 mL | |
| 5 mM | 0.2816 mL | 1.4081 mL | 2.8162 mL | |
| 10 mM | 0.1408 mL | 0.7041 mL | 1.4081 mL |
1、根据实验需要选择合适的溶剂配制储备液 (母液):对于大多数产品,InvivoChem推荐用DMSO配置母液 (比如:5、10、20mM或者10、20、50 mg/mL浓度),个别水溶性高的产品可直接溶于水。产品在DMSO 、水或其他溶剂中的具体溶解度详见上”溶解度 (体外)”部分;
2、如果您找不到您想要的溶解度信息,或者很难将产品溶解在溶液中,请联系我们;
3、建议使用下列计算器进行相关计算(摩尔浓度计算器、稀释计算器、分子量计算器、重组计算器等);
4、母液配好之后,将其分装到常规用量,并储存在-20°C或-80°C,尽量减少反复冻融循环。
计算结果:
工作液浓度: mg/mL;
DMSO母液配制方法: mg 药物溶于 μL DMSO溶液(母液浓度 mg/mL)。如该浓度超过该批次药物DMSO溶解度,请首先与我们联系。
体内配方配制方法:取 μL DMSO母液,加入 μL PEG300,混匀澄清后加入μL Tween 80,混匀澄清后加入 μL ddH2O,混匀澄清。
(1) 请确保溶液澄清之后,再加入下一种溶剂 (助溶剂) 。可利用涡旋、超声或水浴加热等方法助溶;
(2) 一定要按顺序加入溶剂 (助溶剂) 。