| 规格 | 价格 | 库存 | 数量 |
|---|---|---|---|
| 1mg |
|
||
| 5mg |
|
||
| 10mg |
|
||
| 25mg |
|
||
| 50mg |
|
||
| 100mg |
|
||
| 250mg |
|
||
| 500mg |
|
||
| Other Sizes |
|
| 靶点 |
BRD4BD1 (DC50/5h = 500 nM)[1]; DC50: half-maximal degradation concentration, at the half-maximal degradation concentration that degrades 50% of the target protein.
|
|---|---|
| 体外研究 (In Vitro) |
我们发现,dBET6(DC50/5h~10 nM,DC50/5h是指处理5小时后的一半最大降解)、dBET23(DC50/5h~50 nM)和dBET70(DC50/5h~5 nM)对BRD4BD1蛋白水平的影响最为显著,其次是dBET1(8)(DC50/5-500 nM),dBET57(DC50/5k~500 nM)(图3a和补充图4)。对于BRD4BD2,dBET70(DC50/5h~5 nM)的影响最为明显,其次是dBET6(DC50/5-50 nM)、dBET23(DC50/5h>1μM)和dBET1(DC50/5h~1μM)dBET57表现出BRD4BD1的显著退化,对BRD4BD2无效(图3b和补充图4)。因此,细胞活性在很大程度上与观察到的协同因子成正比(补充图3),发现dBET57在生化和细胞测定中对BRD4BD1具有显著的选择性(图2e和图3a,b)。[1]
|
| 酶活实验 |
我们还注意到,具有短连接体的分子,如dBET57 ,将无法在CRBN-dBET23-BRD4BD1结构中观察到的构象中使CRBN和BRD4二聚,因为将E3分子与靶部分桥接需要至少8个碳,而dBET57包含一个2-碳连接体(补充图5c)。因此,我们询问与观察到的结合模式不相容的降解分子,如dBET57或dBET1,是否会以不同的整体构象结合。
为了探索结合的潜在差异,我们进行了突变分析。在CRBN和BRD4BD1中引入了一组单氨基酸点突变,以获得结合的突变特征。除了IMiD结合缺陷(IBD)对照(CRBNP353G W386A)17外,这些CRBN突变之前已被证明与沙利度胺具有相当的亲和力。当比较不同降解剂的突变特征时,我们发现,虽然dBET6和23具有相似的特征(图4a-c和补充图2和5),但dBET1和dBET57的突变特征是不同的(图4d-f和补充图5),这与dBET6/23和dBET五十七的不同结合面是一致的(图4b,e)。这表明,不同的降解分子——取决于接头长度和连接位置——导致CRBN-BRD4复合物形成的不同结合构象。[1]
|
| 参考文献 | |
| 其他信息 |
Heterobifunctional small molecule degraders that induce protein degradation through ligase-mediated ubiquitination have shown considerable promise as a new pharmacological modality. However, we currently lack a detailed understanding of the molecular basis for target recruitment and selectivity, which is critically required to enable rational design of degraders. Here we utilize comprehensive characterization of the ligand dependent CRBN/BRD4 interaction to demonstrate that binding between proteins that have not evolved to interact is plastic. Multiple X-ray crystal structures show that plasticity results in several distinct low energy binding conformations, which are selectively bound by ligands. We demonstrate that computational protein-protein docking can reveal the underlying inter-protein contacts and inform the design of a BRD4 selective degrader that can discriminate between highly homologous BET bromodomains. Our findings that plastic inter-protein contacts confer selectivity for ligand-induced protein dimerization provide a conceptual framework for the development of heterobifunctional ligands.[1]
PROTACs or heterobifunctional degrader molecules (hereafter referred to as degraders) typically comprise an E3 ligase binding scaffold (hereafter E3-moiety), often an analogue of thalidomide, or a ligand to the von Hippel-Lindau tumour suppressor (VHL) protein, attached through a linker to another small molecule (hereafter target-moiety) that binds a target protein of interest. Recruitment of this target protein to the E3 ubiquitin ligase facilitates ubiquitination and subsequent degradation of the target protein. This principle has been successfully applied to several targets including the Bromodomain and Extra Terminal (BET) family (BRD2, BRD3, BRD4), RIPK2, BCR-ABL, FKBP12, BRD9, and ERRα and represents a promising new pharmacologic modality now widely explored in chemical biology and drug discovery. |
| 分子式 |
C34H31CLN8O5S
|
|---|---|
| 分子量 |
699.178544282913
|
| 精确质量 |
698.182
|
| 元素分析 |
C, 58.41; H, 4.47; Cl, 5.07; N, 16.03; O, 11.44; S, 4.59
|
| CAS号 |
1883863-52-2
|
| 相关CAS号 |
1883863-52-2
|
| PubChem CID |
118912822
|
| 外观&性状 |
Typically exists as Light yellow to yellow solids at room temperature
|
| LogP |
3.7
|
| tPSA |
196Ų
|
| 氢键供体(HBD)数目 |
3
|
| 氢键受体(HBA)数目 |
10
|
| 可旋转键数目(RBC) |
8
|
| 重原子数目 |
49
|
| 分子复杂度/Complexity |
1380
|
| 定义原子立体中心数目 |
1
|
| SMILES |
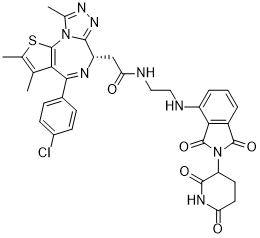
CC1=C(SC2=C1C(=N[C@H](C3=NN=C(N32)C)CC(=O)NCCNC4=CC=CC5=C4C(=O)N(C5=O)C6CCC(=O)NC6=O)C7=CC=C(C=C7)Cl)C
|
| InChi Key |
CZRLOIDJCMKJHE-UXMRNZNESA-N
|
| InChi Code |
InChI=1S/C34H31ClN8O5S/c1-16-17(2)49-34-27(16)29(19-7-9-20(35)10-8-19)38-23(30-41-40-18(3)42(30)34)15-26(45)37-14-13-36-22-6-4-5-21-28(22)33(48)43(32(21)47)24-11-12-25(44)39-31(24)46/h4-10,23-24,36H,11-15H2,1-3H3,(H,37,45)(H,39,44,46)/t23-,24?/m0/s1
|
| 化学名 |
2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethyl)acetamide
|
| 别名 |
dBET57;dBET-57; dBET57; 1883863-52-2; 2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethyl)acetamide; 2-[(9S)-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.02,6]trideca-2(6),4,7,10,12-pentaen-9-yl]-N-[2-[[2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindol-4-yl]amino]ethyl]acetamide; dBET57?; CHEMBL5180012; SCHEMBL17553391; TQP1624; dBET 57
|
| HS Tariff Code |
2934.99.9001
|
| 存储方式 |
Powder -20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month 注意: 本产品在运输和储存过程中需避光。 |
| 运输条件 |
Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs)
|
| 溶解度 (体外实验) |
DMSO : ~250 mg/mL (~357.56 mM)
|
|---|---|
| 溶解度 (体内实验) |
配方 1 中的溶解度: ≥ 2.08 mg/mL (2.97 mM) (饱和度未知) in 10% DMSO + 40% PEG300 +5% Tween-80 + 45% Saline (这些助溶剂从左到右依次添加,逐一添加), 澄清溶液。
例如,若需制备1 mL的工作液,可将100 μL 20.8 mg/mL澄清DMSO储备液加入400 μL PEG300中,混匀;然后向上述溶液中加入50 μL Tween-80+,混匀;加入450 μL生理盐水定容至1 mL。 *生理盐水的制备:将 0.9 g 氯化钠溶解在 100 mL ddH₂O中,得到澄清溶液。 请根据您的实验动物和给药方式选择适当的溶解配方/方案: 1、请先配制澄清的储备液(如:用DMSO配置50 或 100 mg/mL母液(储备液)); 2、取适量母液,按从左到右的顺序依次添加助溶剂,澄清后再加入下一助溶剂。以 下列配方为例说明 (注意此配方只用于说明,并不一定代表此产品 的实际溶解配方): 10% DMSO → 40% PEG300 → 5% Tween-80 → 45% ddH2O (或 saline); 假设最终工作液的体积为 1 mL, 浓度为5 mg/mL: 取 100 μL 50 mg/mL 的澄清 DMSO 储备液加到 400 μL PEG300 中,混合均匀/澄清;向上述体系中加入50 μL Tween-80,混合均匀/澄清;然后继续加入450 μL ddH2O (或 saline)定容至 1 mL; 3、溶剂前显示的百分比是指该溶剂在最终溶液/工作液中的体积所占比例; 4、 如产品在配制过程中出现沉淀/析出,可通过加热(≤50℃)或超声的方式助溶; 5、为保证最佳实验结果,工作液请现配现用! 6、如不确定怎么将母液配置成体内动物实验的工作液,请查看说明书或联系我们; 7、 以上所有助溶剂都可在 Invivochem.cn网站购买。 |
| 制备储备液 | 1 mg | 5 mg | 10 mg | |
| 1 mM | 1.4302 mL | 7.1512 mL | 14.3025 mL | |
| 5 mM | 0.2860 mL | 1.4302 mL | 2.8605 mL | |
| 10 mM | 0.1430 mL | 0.7151 mL | 1.4302 mL |
1、根据实验需要选择合适的溶剂配制储备液 (母液):对于大多数产品,InvivoChem推荐用DMSO配置母液 (比如:5、10、20mM或者10、20、50 mg/mL浓度),个别水溶性高的产品可直接溶于水。产品在DMSO 、水或其他溶剂中的具体溶解度详见上”溶解度 (体外)”部分;
2、如果您找不到您想要的溶解度信息,或者很难将产品溶解在溶液中,请联系我们;
3、建议使用下列计算器进行相关计算(摩尔浓度计算器、稀释计算器、分子量计算器、重组计算器等);
4、母液配好之后,将其分装到常规用量,并储存在-20°C或-80°C,尽量减少反复冻融循环。
计算结果:
工作液浓度: mg/mL;
DMSO母液配制方法: mg 药物溶于 μL DMSO溶液(母液浓度 mg/mL)。如该浓度超过该批次药物DMSO溶解度,请首先与我们联系。
体内配方配制方法:取 μL DMSO母液,加入 μL PEG300,混匀澄清后加入μL Tween 80,混匀澄清后加入 μL ddH2O,混匀澄清。
(1) 请确保溶液澄清之后,再加入下一种溶剂 (助溶剂) 。可利用涡旋、超声或水浴加热等方法助溶;
(2) 一定要按顺序加入溶剂 (助溶剂) 。
|
|
|